Contents
Insert or bind?
This is the first in a series of planned posts, sharing some R tips and tricks. I hope to cover topics which are not easily found elsewhere. This post has to do with loops in R. There are two ways to save values when looping:
1. You can predefine a vector and fill it, or
2. you can recursively bind the values.
Which one is faster?
The package microbenchmark provides infrastructure to accurately measure and compare the execution time of R expressions. You can use it when the code is too fast to be timed using a stopwatch.
We can check different vector lengths. It turns out that the preferred method depends on the vector size.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 |
# Option one - define y and fill it in fun_insert <- function(k= 100) { x <- runif(k) y <- NULL for (i in 1:k){ y[i] <- x[i] } y } # Option two - define y and bind each value fun_bind <- function(k= 100) { x <- runif(k) y <- NULL for (i in 1:k){ y <- c(y, x[i]) } y } library(microbenchmark) # install if you don't have it yet kk <- c(50, 100, 200, 500, 1000, 2000) lkk <- length(kk) run_times <- matrix(ncol= 2, nrow= lkk) for (i in 1:lkk) { tmpp <- microbenchmark( fun_insert(k= kk[i]), fun_bind(k= kk[i]), unit= "us" ) run_times[i,] <- summary(tmpp)$med } |
Here are the results:
| Vector length | Insert | Bind |
|---|---|---|
| 50 | 73.5 | 35.9 |
| 100 | 120.3 | 70.0 |
| 200 | 280.1 | 192.8 |
| 500 | 694.9 | 538.5 |
| 1000 | 1636.5 | 1614.1 |
| 2000 | 7459.0 | 8584.6 |
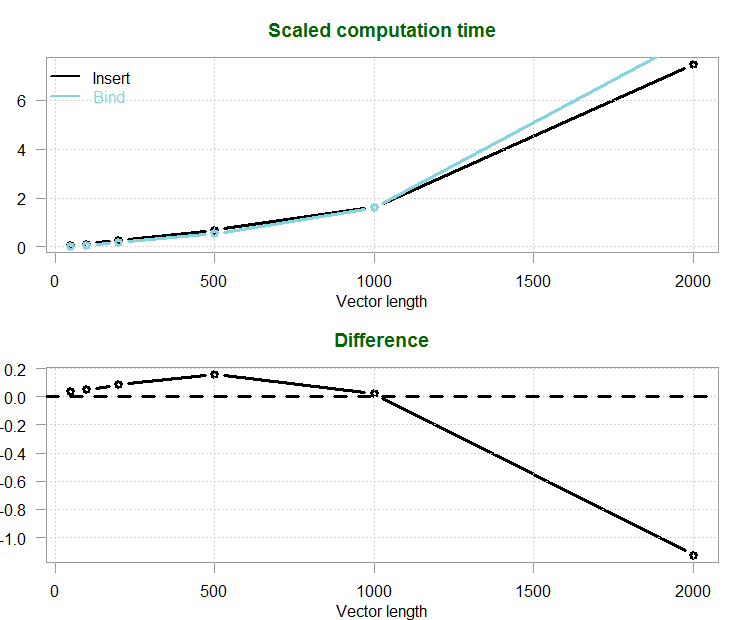
I actually wonder why the difference is not a monotonic function of the vector length. I guess R makes some choices on its own somewhere. It doesn’t matter, the conclusion is clear enough: for short vectors use the bind method, for longer vector fill in the values. This is particularly relevant when we perform Monte Carlo simulations or bootstrapping, where the sample is not very big on the one hand, but we would like to create many simulations on the other hand. Binding instead of filling can save time in those situations. When you have long vectors, stick with filling-in the values.
Update – preallocation
Rightfully mentioned by an attentive reader, we can improve performance by preallocating. If we know how many loops we need, we can share this info with the machine and be rewarded with lessened computational time. Simply replace the line
y <- NULL
with
y <- rep(NA, k)
and rerun the code. Here is the resulting table:
| Insert | Bind | |
|---|---|---|
| 50 | 59.8 | 50.0 |
| 100 | 99.2 | 99.6 |
| 200 | 190.3 | 316.8 |
| 500 | 502.6 | 1945.0 |
| 1000 | 881.2 | 3472.9 |
| 2000 | 1850.2 | 12717.0 |
As you can see it is much faster. We can use it when we know beforehand how many loops we have (a while loop is a contrasting example).
Fast subsetting
The reader also provided a reference added below (Hadley of course). What I found especially nice there is the following. I usually use indexing for subsetting, e.g. run_times[1,2] to extract the element in the first row and second column. Not to repeat the analysis, you can find it in the link below. It appears that the function .subseting2 impressively tamps down computational time. So for extracting single value we can use
.subset2(run_times, i= 1, j= 2).
We can discuss readability, but no doubt it is much faster. Worth having in your back pocket.
References
Olaf Mersmann (2015). microbenchmark: Accurate Timing Functions. R package version 1.4-2.1. https://CRAN.R-project.org/package=microbenchmark
Advanced R (Performance)
Code Complete: A Practical Handbook of Software Construction










Perhaps a third way — pre-allocate the target vector.
fun_alloc <- function(k=100) {
x <- runif(k)
y <- rep(NA,k)
for ( i in 1:k ) {
y[i] <- x[i]
}
y
}
For me this produces near-linear and much faster times. Another angle on the test is to eliminate the 'for' construct and use R's apply family. For me this also produced much faster times.